You are here
Lemer Invertebrate Genomics Lab
Lemer Invertebrate Genomics Lab

About

Sarah Lemer, PhD (CV)
Assistant Professor of Marine Invertebrate Genomics
University of Guam Marine Laboratory
UOG Station, Mangilao, Guam 96923 USA
sarah.lemer@gmail.com
Google Scholar Profile / ResearchGate Profile
Other Affiliation:
Research Associate
Department of Organismic & Evolutionary Biology
Museum of Comparative Zoology
Harvard University
Research Interests
 Research in the Lemer lab focuses on understanding how biodiversity arises and is
maintained in marine invertebrates, at the species, population and individual levels,
in the context of a changing environment.
Research in the Lemer lab focuses on understanding how biodiversity arises and is
maintained in marine invertebrates, at the species, population and individual levels,
in the context of a changing environment.
Our work primarily focused on diverse mollusc groups, but we also explore the wider realm of marine invertebrates to address our research questions (We even sometimes venture into the vertebrate world!).
In the Lemer lab we mainly use a combination of field work, experiments in controlled environments, Next Generation Sequencing approaches (RNA-Seq, RAD-Seq, Tag-Seq and Genome sequencing) and bioinformatics to answer questions about population genetics, phylogeny and gene expression in various taxa such as Annelids, Bivalves, Cephalopods and Scleractinian corals.
The Lemer Lab stands for justice and equal treatment of people regardless of race, origin, gender or sexual orientation. As such we promote inclusive, anti-racist, anti-sexist and anti-discriminatory practices. The PI is committed to actively work towards advancing women, people of color and pacific islanders in the lab and the classroom with opportunities such as science education, lab experience, conference attendance and outreach. We welcome new students and postdocs who share our values and are passionate about invertebrate evolutionary genomics, contact me: sarah.lemer@gmail.com
I am part of the NSF funded Guam Ecosystems Collaboratorium. Learn more about what we do here: Guam EPSCoR
Current Projects

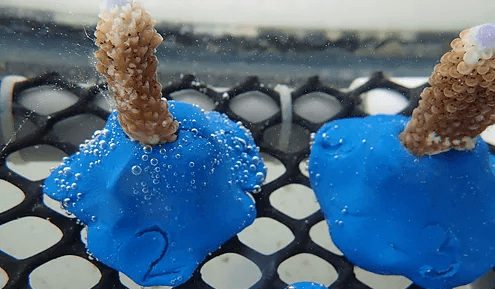
Bleaching Resistance
We are using in aquaria experiments to reproduce the extreme environmental conditions that lead to coral bleaching. The goal is to identify and characterize the molecular processes that allow some corals to adapt to and survive extreme environmental conditions when others usually bleach and/or die.
Bivalve Phylogenomics
Phylogenetic relationships among bivalves still show areas of uncertainty, especially within the Imparidentia group. We sequence and analyze bivalve transcriptomes and genomes to shed light on the phylogeny of this diverse group of molluscs.
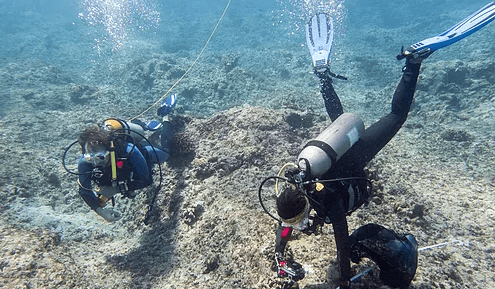
Coral and Other Invertebrate Population Genomics
Species and population resilience to changing environmental conditions depend on genetic diversity and connectivity. Generally, genetic diversity and connectivity of reef-building and reef-associated taxa (e.g., corals, crustaceans, algae, mollusks) remain poorly understood in the western Pacific, an issue we will address. Being the arguably most diverse reef ecosystem within US jurisdiction, Guam and the CNMI represent a unique opportunity to study reef connectivity and resilience in a changing climate. Recovery of benthic communities following disturbance relies on connectivity but is also dependent on ecological processes like the recruitment of larvae from nearby populations. We are exploring the genetic connectivity, diversity and structure of multiple coral species in the Micronesia using a RAD-Seq or Shallow Whole Genome Sequencing approaches.
Studied species: Trapezia crabs, Tridacna clams, Drupella snails, COTs, Acropora surculosa, Leptastrea purpurea, etc...
Micronesian Reef Fish Phylogeography
In Micronesia, the fishing industry is a local source for food and economic development, yet movement of fish between Micronesian islands remains unknown. We hope to better understand genetic structure and diversity patterns of the fish families Acanthuridae, Lutjanidae, Scaridae and Lethrinidae across islands.
Lemer Lab Team and Associates!

Postdocs

Carlos Leiva
Research: Population genomics and Biogeography of the small giant clam Tridacna maxima
Current position: Postdoctoral Researcher

Heloise Rouzé
Research: Characterizing Gene Expression and Microbiome Patterns of Early Stress Response in Acropora corals
Current position: Postdoctoral Researcher
Graduate students

Kenzie Pollard
Graduate Research Assistant
Research:Phylogeography and Population Genomics of Trapezia bidentata in the Indo-Pacific

Olivia Barry
Graduate student Assistant
Research: Population Genomics of the heat resistant coral Leptastrea purpurea on Guam

Monica Salas
Graduate student Assistant
Research: Population Genetics of the crown-of-thorn sea star Acanthaster planci through time in the Mariana Archipelago
Undergraduates

Laura Caser
Biology undergraduate
Research: Identifying species of corallivore Drupella snails on Guam

Lynn Galang
Biology Undergraduate
Alumni
Postdocs
- Pierre-Louis Stenger: Epigenetics and Gene Expression patterns of Coral Acclimation to Heat Stress
Graduates
- Constance Sartor: Transcriptomic Signatures of Acclimation in 'susceptible' Acropora surculosa and 'resistant' Porites rus
- Victoria Moscato: Characterizing the Role of Color in Heat Stress Resistance in the Scleractinian coral Acropora surculosa
Undergraduates
- Nikko Galanto: Effect of heat stress on the reproduction of Leptastrea purpurea
- Anela Duenas: Sediment stress on Acropora corals
- Amhiya Cacapit: Sediment stress on Acropora corals
- Mikel Lizama: Effect of heat stress on the reproduction of Leptastrea purpurea
- Charles Hambley: Characterisatoin of abiotic conditions triggering glowing phenotypes in corals.
- Joanna Panaguiton: Effect of heat stress on the reproduction on Leptastrea sp. corals.
- Jasmin Rotan: Micronesian Reef Fish Phylogeography
Selected Publications
Google Scholar Profile / ResearchGate Profile
2021
- YH. Phua*, MC. Roy, S. Lemer, F. Husnik, KC. Wakeman. 2021. Diversity and toxicity of Pacific strains of the benthic dinoflagellate Coolia (Dinophyceae), with a look at the Coolia canariensis species complex. Harmful Algae. 109: 102-120
- V. Fabian**, P. Houk, S. Lemer. 2021. Resolving the Phylogenetic Relationship of Micronesian Emperor Fishes. Molecular Phylogenetics and Evolution. 162 (2021):107207
- J. Moles, TJ. Cunha, S. Lemer, D. Combosch, G. Giribet. 2021. Tightening the girdle : Phylotranscriptomics of Polyplacophora. Journal of Molluscan Studies 84(2):eyab019
- P. Houk, S. Lemer, D. Hernandez, J. Cuetos-Bueno. 2021. Evolutionary management of coral-reef fisheries using phylogenies to predict density dependance. Ecological applications :e02409
- Fifer*, B. Bentlage, S. Lemer, A. Fujimura, M. Sweet, L. Raymundo. 2021. Going with the flow: Corals in high-flow environments can beat the heat. Molecular Ecology https://doi.org/10.1111/mec.15869
2020
- Li; S. Lemer; L. Kirkendale; R. Bieler; C. Cavanaugh; G. Giribet. 2020. Shedding light: A phylotranscriptomic perspective illuminates the origin of photosymbiosis in marine bivalves. BMC Evolutionary Biology, 20, 1-15
2019
- Laumer, R. Fernandez, S. Lemer, D. Combosch, K. Kocot, A. Riesgo, S. Andrade, W. Sterrer, M. Sorensen, G. Giribet. 2019. Revisiting metazoan phylogeny with genomic sampling oof all phyla. Proceedings of the Royal Society B: 286
- S. Lemer, R. Bieler, G. Giribet. 2019. Resolving the relationships of clams and cockles: dense transcriptome sampling drastically improves the bivalve tree of life. Proceedings of the Royal Society B: 283
- T. Cunha, S. Lemer, P. Bouchet, Y. Kano, G. Giribet. 2019. Putting keyhole limpets on the map: Phylogeny and biogeography of the global marine family Fissurellidae (Vetigastropoda, Mollusca). Molecular Phylogenetics and Evolution; 135:249-269
2017
- D. Combosch*, S. Lemer*, N. Landman, P. Ward, G. Giribet. 2017. Genomic signatures of evolution in the living fossil Nautilus. Molecular Ecology, 26:5923-5938. * Shared first authorship
- PUPA. Gilbert, KD. Bergmann, CE. Myers, RT. DeVol, CY. Sun, AZ. Blonsky, J. Zhao, EA. Karan, E. Tamre, N. Tamura, MA. Marcus, AJ. Giuffre, S. Lemer, G. Giribet, JE. Eiler, AH. Knoll. 2017. Nacre tablet thickness records formation temperature in modern and fossil shells. Earth and Planetary Science Letters; 460:281-292
2016
- D. Combosch; TM. Collins; EA, Glover; DL. Graf; EM. Harper; JM. Healy; GY. Kawauchi; S. Lemer; E. McIntyre; EE. Strong; JD. Taylor; JD. Zardus; PM. Mikkelsen; G. Giribet, R. Bieler. 2017. A family-level Tree of Life for bivalves based on a Sanger-sequencing approach. Molecular Phylogenetics and Evolution, 107: 191-208
- S. Lemer, D. Combosch, D. Dumale, F. Sotto, V. Soliman, G. Giribet. 2016. The family Pinnidae (Mollusca, Bivalvia) in the Philippine archipelago: observations on its distribution and phylogeography. The Nautilus, 130:4.
- S. Lemer, V. González, R. Bieler, G. Giribet,. 2016. Cementing mussels to oysters in the pteriomorphian tree: phylogenomic approach. Proceedings of the Royal Society B, 283:1833
2015
- S. Lemer, D. Saulnier, Y. Gueguen, S. Planes,. 2015. Identification of genes associated with shell color in the black-lipped pearl oyster, Pinctada magaritifera. BMC Genomics, 16:568
- S. Lemer, GY. Kawauchi, CS Andrade, MJ Boyle, G. Giribet,. 2015. Re-evaluating the phylogeny of Sipuncula through transcriptomics. Molecular Phylogenetics and Evolution, 83: 174-183
- R. Fernandez*, S. Lemer*, G. Giribet,. 2015. Comparative phylogeography and population genetic structure of three widespread mollusc species in the Mediterranean and near Atlantic. Marine Ecology, 36: 701-715. * Shared first authorship
2014
- S. Lemer & G. Giribet,. 2014. Occurrence of a Bivalve-inhabiting marine hydrozoan (Hydrozoa, Hydroidolina, Leptothecata) in the amber pen-shell Pinna carnea Gmelin, 1791 (Bivalvia, Pteriomorphia, Pinnidae) from Bocas del Toro. Journal of Molluscan Studies, 80: 464-468
- G. Giribet, S. Lemer,. 2014. On the occurrence of Tuleocaris neglecta Chace, 1969 (Decapoda, Palaemonidae, Pontoniinae) in Echinometra lucunter (Linnaeus, 1758) (Echinodermata, Echinoidea, Echinometridae) in the archipelgos of Bocas del Toro, Panama. Crustaceana, 87: 634-638
- S. Lemer & S. Planes,. 2014. Effects of habitat fragmentation on the genetic structure and connectivity of the black‑lipped pearl oyster Pinctada margaritifera populations in French Polynesia. Marine Biology, 161: 2035-2049
- S. Lemer, B Buge, A. Bemis, G. Giribet. 2014. First molecular phylogeny of the circumtropical bivalve family Pinnidae (Mollusca, Bivalvia): evidence for high levels of cryptic species diversity. Molecular Phylogenetics and Evolution, 75: 11-23
2012
- S. Lemer & S. Planes,. 2012. Translocation of wild populations: conservation implications for the genetic diversity of the black-lipped pearl oyster Pinctada margaritifera. Molecular Ecology, 21: 2949-2962
- A.C. Stier, M.A. Gil, C.S. McKeon, S. Lemer, M. Leray, S.C. Mills, C.W. Osenberg,. 2012. Housekeeping mutualisms: do more symbionts facilitate host performance? PLoS ONE 7(4):e32079. doi:10.1371/journal.pone.0032079
2011
- S. Lemer, E. Rochel, S. Planes,. 2011. Correction method for null alleles in species with variable microsatellite flanking regions, a case study of the black-lipped pearl oyster Pinctada margaritifera. Journal of Heredity, 102 (2) 243-246
- S. Planes & S. Lemer,. 2011. Individual-based analysis opens new insights into understanding population structure and animal behaviour. Molecular Ecology, 20: 187-189
2007
- P. Borsa, S. Lemer, D. Aurelle,. 2007. Patterns of lineage diversification in rabbitfishes. Molecular Phylogenetics and Evolution, 44: 427-435.
2006
- S. Lemer , D. Aurelle, L. Vigliola, J.D. Durand, P. Borsa,. 2006. Cytochrome b barcoding, molecular systematics, and geographic differentiation in rabbitfishes (Siganidae). Comptes Rendus Biologies, 330 (1): 86-94.
Teaching: Biology Graduate Program
BI–546 MARINE INVERTEBRATES
FALL
4 credit hours
BI546 is a 4-credit graduate level course that will introduce you to the major (and not-so-major) marine invertebrate phyla. Lectures will address topics of higher-level phylogeny, anatomical and morphological diversity, life history, ecology, anatomy and physiology of each group. Laboratories will provide hands-on experience with representatives of the groups we cover in lecture. We will go to field sites as well, so that you can get experience identifying invertebrates in the field. The class will meet for two lectures per week on Tuesdays and Thursdays 2:00-3:20 PM and one laboratory on Friday 1:00-3:50 PM.
BI–691 SEMINAR: INTRODUCTION TO BIOGEOGRAPY
SPRING
1 credit hour
BI691 is a 1-credit graduate level course that will introduce you to the ecological and historical processes that affect distributions of organisms. The following topics are adressed through scientific paper presentation and discussion: plate tectonics and earth history, vicariance and dispersal, endemism, conservation biogepgraphy, latitudinal gradients in species richness, and the theory of island biogeography.